The Omixon HLA Genotyping Portal is now live
The Omixon Genotyping Portal (OGP) consumes fastq.gz files for whole-genome or whole-exome sequenced (WGS/WES) subjects, and generates HLA-A, -C, -B, -DRB1, -DRB3, -DRB4, -DRB5, -DQA1, -DQB1, -DPA1 and -DPB1 genotypes for each subject. Because WGS/WES fastq.gz files can be very large, only fastq.gz files containing HLA-gene specific reads can be uploaded to the OGP. The wgsHLAfiltR R package can be used to generate HLA gene-specific fastq.gz files.
The OGP can only be applied to subjects with origin identifiers that have been previously loaded into the HCDB under a specific Project Name. To create a HCDB account and/or load origin identifiers and clinical data for subjects, visit https://database-hlacovid19.org.
The OGP relies on Omixon’s CLI Explore software to generate HLA genotypes using IPD-IMGT/HLA Database release version 3.39.0 sequence alignments. CLI Explore performs HLA genotyping on the basis of HLA exon sequence information. Given this, the OGP generates HLA genotypes with fourth-field ambiguity, reported in GL String format (dx.doi.org/10.1111/tan.12150).
The OGP can consume paired or unpaired fastq.gz files, and each fastq.gz pair or single fastq.gz file must be associated with a specific origin identifier. To structure the submission of this information, the OGP consumes a three-column “User Information” file written in CSV format (shown below).
Example OGP User Information file
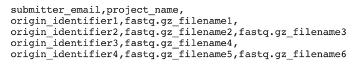
The first line of the User Information file should contain the email associated with the submitter’s HCDB account, and the Project Name associated with the sequenced subjects, separated by a comma, and followed by a comma.
Each subsequent line of the User Information file should contain the origin identifier of each subject and the name of the unpaired fastq.gz file, or the names of the paired fastq.gz files, associated with that subject. These elements must be separated by commas, and each row of the file must include two commas. If only one fastq.gz filename is provided, it should be followed by a comma. As noted above, these origin identifiers must have been loaded into the HCDB before using the OGP.
All of the fastq.gz files specified in the User Information file should be located in the same directory (or folder) on your system. To minimize upload time, exclude all other fastq.gz files from that directory.
To access the OGP, go to https://database-hlacovid19.org/shiny in your browser, and click Omixon-Genotyping-Portal/. Once connected to the OGP, click the ‘Browse…’ button under User Info .csv, and select the User Information file. Once that file has been uploaded, click the ‘Browse…’ button under FASTQ.GZ files and select the directory containing the fastq.gz files. Click the box indicating that your fastq.gz files have been filtered to include only HLA-gene reads. Once those files have been uploaded (which can take a few minutes), click the Genotype! button to start the genotyping process.
